Introduction
Artificial chromosomes (ACs) are high-capacity vectors that possess essential chromosomal elements or mimic natural chromosomes: centromeres, telomeres, and specific sequences necessary for autonomous self-replication and stable maintenance within cells (Singh et al., 2020). As high-capacity vectors, ACs overcome the size limitations inherent in conventional vectors. This article provides a comprehensive introduction to Yeast Artificial Chromosomes (YACs), the construction of YACs, applications, limitations, and a comparison to Bacterial Artificial Chromosomes (BACs).
The main functions of a telomere are to maintain chromosomal stability and prevent chromosomal degradation, essential to eukaryotic genomic stability and the longevity of cellular information.
#
Centromeres are segments of highly repetitive DNA essential for the proper control of chromosome distribution during cell division.
#
Autonomously Replicating Sequences (ARS) are specific DNA elements that serve as independent origins of replication, functioning separately from the primary chromosomal replication origins. Artificial chromosomes serve as shuttle vectors that can replicate and be maintained across multiple host organisms, such as eukaryotic and prokaryotic cells. This compatibility with both hosts arises from incorporating distinct replication origins and selective markers tailored to each host system. This enables genetic manipulation in one organism (E-coli) and expression or study in another (yeast).
Yeast Artificial Chromosomes (YACs)
Yeast artificial chromosome (YAC) is a shuttle plasmid vector allowing the cloning within yeast cells of large fragments of foreign genomic DNA, larger than 100Kbp up to 3000 kbp in size. YACs are useful for the physical mapping of complex genomes and for the cloning of large genes (Bajpai, 2013). It mimics a natural chromosome in yeast cells and includes essential elements such as ARS, CEN, and TEL. YAC vectors contain the following essential elements:
- The ARS1 (Autonomously Replicating Sequence) and CEN4 (Centromere on Chromosome 4) elements are naturally adjacent to the TRP1 gene on yeast chromosome IV. ARS1 provides the sequence required for autonomous replication, while CEN4 confers centromere function. Centromeres are essential for proper chromosome segregation during cell division.
- The Tetrahymena telomere (TEL) sequences protect the ends of the YAC from damage. The ends of the linear YAC incorporate telomeric sequences derived from the termini of Tetrahymena macronuclear ribosomal DNA (rDNA) molecules to mimic natural chromosomal termini.
- The genes used for YAC selection in yeast are TRP1 and URA3. These genes allow for the selection of transformants that have acquired both chromosome arms from the vector. The construction process discards the HIS3 gene. The YAC is transformed into a host yeast cell defective in the biosynthetic pathways for tryptophan and uracil. They identify transformants by observing their ability to complement these nutritional defects and grow on media lacking these amino acids.
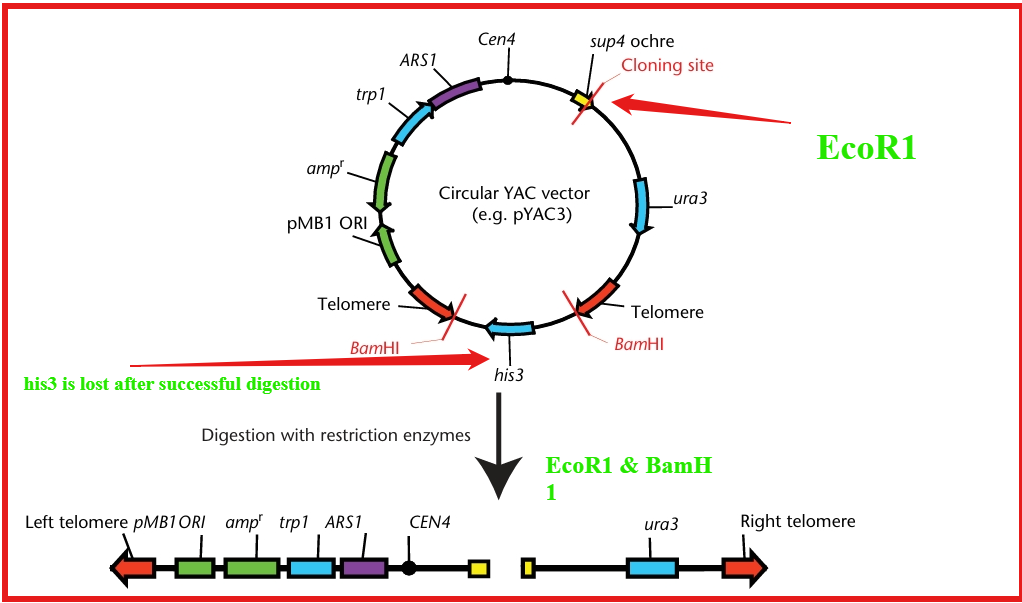
Construction of YACs
- After purifying the plasmid DNA, we cleave the YAC with the restriction enzyme BamH1 to generate a long linear fragment with telomere sequences at the ends in pYAC3. The telomeric sequence flanks the his3 gene used as a negative marker used to select uncut pYAC molecules.
- Digesting with EcoR1 cuts the multiple cloning site(MCS) within the sup4 gene. This results in two linear fragments denoted as left and right arms each having EcoR1(sup4 end) and BamH1(telomere end) cleavage.
- The left arm contains a selectable marker (TRP1) adjacent to the autonomous replication sequence (ARS1) and a centromere (CEN4). The right arm contains a second selectable marker (URA3)
- An appropriate restriction endonuclease digests the desired genome to obtain properly sized DNA fragments. These fragments are then ligated into the cloning site of the linear YAC vector arms. This inactivates the suppressor tRNA gene SUP4 which normally expresses tRNATyr.
- When introduced into a yeast cell with a mutation in the ADE2 gene it blocks the adenine biosynthetic pathway and causes an intermediate to accumulate in the vacuole.
- The inactivation of SUP4 results in the formation of red yeast colonies due to the accumulated intermediate. In contrast, the expression of the functional SUP4 gene leads to white colonies.
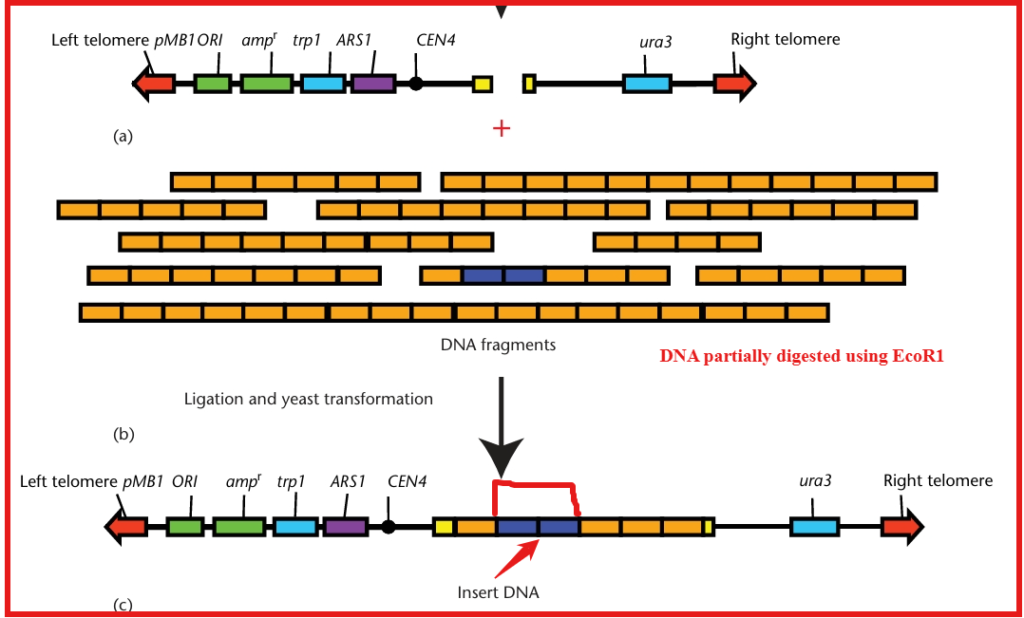
Selection of transformed recombinant YACs
- The yeast strain, which has defects in its chromosomal copies of the URA3, TRP1, and ADE2 genes, receives the recombinant YACs through transformation. We identify transformants as the red colonies that grow on media lacking both uracil and tryptophan.
- This ensures that the cell has received an artificial chromosome with both telomeres (because of the complementation of the two nutritional mutations). The artificial chromosome contains insert DNA (because the cell is red due to the ADE2 gene defect).
Selection of transformed recombinant YACs: Transform into ade−, ura−, trp− yeast and select for red, URA+, TRP+ colonies Application of YACs
| Application | Description | Reference |
|---|---|---|
| Genome Mapping and Sequencing | YACs have been widely used for the construction of physical maps and the sequencing of large genomic regions in various organisms, including humans. | Green and Olson, 1990; Shizuya et al., 1992 |
| Exploring Genomic Instability | One of the most important uses of YACs is the exploitation of their behavior as endogenous chromosomes to develop assays in budding yeast to identify mutants with enhanced rates of gross chromosomal rearrangements (GCRs). | Putnam et al., 2016; Stirling et al., 2011 |
| Functional Genomics | YACs can be used to study the function of genes and regulatory elements by introducing them into cells or organisms and observing the phenotypic effects. YAC clones have been used as hybridization probes for the screening of complementary DNA (cDNA) libraries. | Schedl et al., 1993; Peterson et al., 1997 |
| Transgenic Studies | YACs can be used to generate transgenic animals or plants by introducing large genomic regions, allowing for the study of complex traits and the regulation of gene expression in their native genomic context. | Schedl et al., 1993; Mullins et al., 1997 |
| Disease Gene Identification | YACs have been instrumental in the identification and cloning of disease-related genes, particularly for conditions associated with large, complex genomic regions. YACs allow positional cloning of disease genes and the preparation of DNA probes for diagnostic purposes. | Rossiter et al., 1991; Collins, 1995 |
| Heterologous Gene Expression | YACs can be used to express large, complex genes or gene clusters in a eukaryotic host, such as yeast, facilitating the production of therapeutic proteins or the study of metabolic pathways. | Hieter and Boguski, 1997; Kouprina and Larionov, 2016 |
| Metagenomics and Environmental Studies | YACs have been employed in metagenomic studies to clone and analyze large genomic fragments from uncultured microorganisms, expanding our understanding of microbial diversity and function in various environments. | Rondon et al., 2000; Gabor et al., 2004 |
| Fluorescence in situ Hybridization (FISH) | YACs can be labeled with fluorochromes and applied in FISH experiments for gene mapping, chromosome characterization, and studies on interphase architecture and karyotypic evolution. | Liehr, 2006 |
Limitation of YACs
| Difficulty | Description |
|---|---|
| Fragility and Rearrangements | Very large DNA molecules in YACs are prone to breakage, leading to problems of rearrangement. It is estimated that 10-60% of YAC clones in genomic libraries are chimeric, meaning regions from different parts of the genome are joined in a single YAC clone. |
| Instability and Deletions | YAC clones tend to be unstable, with their foreign DNA inserts often being deleted. The insertion of repetitive DNA sequences can increase the recombination frequency within the YAC, further contributing to instability. |
| Loss during Mitotic Growth | There is a high rate of loss of the entire YAC during mitotic growth in yeast cells. |
| Difficulty in Separation | It is challenging to separate the YAC from the other host chromosomes due to their similar size. This separation requires sophisticated techniques like pulsed-field gel electrophoresis (PFGE). |
| Low DNA Yield | The yield of DNA obtained when isolating the YAC from yeast cells is generally not high. |
Difference between YAC and BAC Vectors
The key differences between YAC (Yeast Artificial Chromosome) and BAC (Bacterial Artificial Chromosome) vectors.
| Feature | YAC | BAC |
|---|---|---|
| Host Organism | Yeast (Saccharomyces cerevisiae) | Escherichia coli |
| Insert Size Capacity | Very large, up to 2 Mb | Large, up to 300 kb |
| Replication Mechanism | Autonomous replication using yeast ARS sequences | Controlled replication using the F-factor origin of replication |
| Structural Features | Contains yeast centromere (CEN), autonomously replicating sequence (ARS), and telomeres | Contains bacterial ori, selectable marker, and F-factor sequences (repE, parA, parB and parC) |
| Stability | Very large DNA molecules are very fragile and prone to breakage, leading to problems of rearrangement. | BAC appears to be very stable and to be less prone to rearrangements and deletions when maintained in a recombination defective E. coli host cell. |
| Transformation Efficiency | Lower than BACs, typically around 10^4 transformants per μg DNA | Higher than YACs, up to 10^6 transformants per μg DNA |
| Applications | Ideal for cloning and studying very large genomic regions | Suitable for a wide range of applications, including genome sequencing and functional genomics |
References
- Burke, D T et al. “Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors.” Science (New York, N.Y.) vol. 236,4803 (1987): 806-12. doi:10.1126/science.3033825
- Singh, P., Mondal, S., & Singh, R. L. (2020). Introduction. In Elsevier eBooks (pp. 1–12). https://doi.org/10.1016/b978-0-12-820595-2.00001-1.
- Bajpai, B. (2013). High capacity vectors. In Springer eBooks (pp. 1–10). https://doi.org/10.1007/978-81-322-1554-7_1.
- Reece, R. J. (2004). Analysis of genes and genomes. Wiley.
- Arnak, R., Bruschi, C. V., & Tosato, V. (2012). Yeast artificial chromosomes. Encyclopedia of Life Sciences. doi: 10.1002/9780470015902.a0000379.pub3
- Liehr T. (2006). Application of yeast artificial chromosomes in fluorescence in situ hybridization. Methods in molecular biology (Clifton, N.J.), 349, 175–186. https://doi.org/10.1385/1-59745-158-4:175.


