Summary
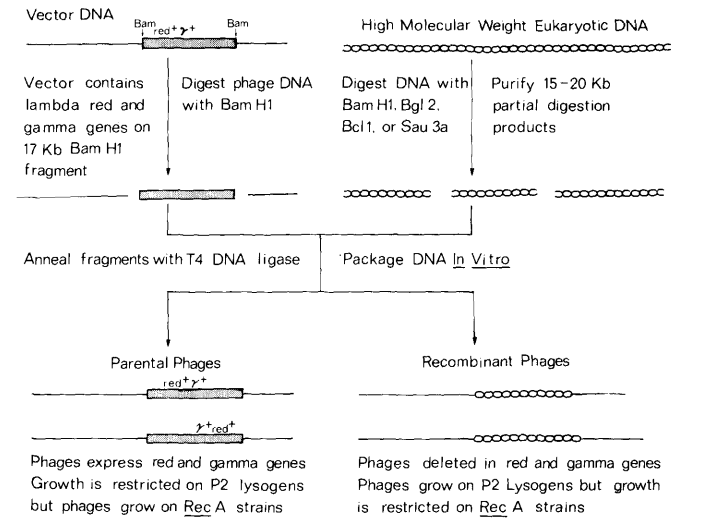
The Lambda EMBL3 and EMBL4 vectors are high-capacity, phage lambda-derived replacement vectors developed by the EMBL* representing a significant advancement in genomic library construction and large-insert cloning. Karn et al. (1983) derived the EMBL3 and EMBL4 vectors from lambda 1059, modifying the symmetrical polylinker sequences from plasmid pUC2 [3*] to flank a central 14 kb [2] stuffer fragment that contains the red and gam genes, which confer sensitivity to interference by the P2 lysogen [4]. Researchers replace the stuffer DNA with DNA inserts of approximately 8 to 23 kb [2], without affecting phage viability, which facilitates the selection of phenotypes sensitive to P2 interference (Spi-) [2].
Introduction
Lambda EMBL3 and EMBL4 vectors feature modified symmetrical polylinker sequences from plasmid pUC2 (Vieira & Messing, 1982), flanking a central stuffer fragment containing red+ and gam+ genes conferring sensitivity to interference by the P2 lysogen. During cloning, the stuffer DNA is replaced with DNA insert without affecting phage viability enabling selective propagation of recombinant phage. The Lambda EMBL3 and EMBL4 vector systems have emerged as powerful tools in genomic library construction, offering high cloning capacity and an ingenious selection method based on the Spi- phenotype.
The Spi-negative selection method capitalizes on the fact that lambda phages with active red and gam genes cannot grow on bacteriophage P2 lysogenic host strains. When the stuffer region is replaced by an insert, the recombinant phage becomes red-/gam-, enabling growth on P2 lysogenic strains such as XL1-Blue MRA (P2) (Agilent Technologies, 2008). This selective plating method ensures that only recombinant phages propagate, providing an efficient means of isolating desired clones.
This review provides a comprehensive analysis of the Lambda EMBL3 and EMBL4 vector selection methods using the Spi-phenotype mechanism. It explores the design principles, the molecular basis of selection, and practical applications of these vectors in genomic research. Additionally, recent advancements in the EMBL vector series, including the Lambda GEM11 and GEM12 derivatives, are discussed, highlighting their impact on functional genomics and systems biology
Principle of the Method
Lambda EMBL3 and EMBL4 vectors are derived from lambda 1059 (Karn et al., 1983), by replacing the BamHI cloning sites of EMBL2 with modified symmetrical polylinker sequences from plasmid pUC2 [2]. Both vectors feature polylinker sequences, or multiple cloning sites (MCS), flanking a central stuffer fragment. This stuffer region contains the red+ and gam+ genes, which confer sensitivity to interference by the P2 lysogen. During cloning, the stuffer DNA is replaced with DNA inserts of approximately 8 to 23 kb (Frischauf et al., 1989) without affecting phage viability facilitating the selection of the Spi- (sensitive to P2 interference) phenotype.
The Spi-negative selection method capitalizes on the fact that lambda phages with active red and gam genes cannot grow on bacteriophage P2 lysogenic host strains. The stuffer can be excised using flanking SalI (EMBL3) or EcoRI (EMBL4) sites. The primary distinction between these vectors lies in the orientation of the restriction sites within their respective MCS.
recA+ strains
Lambda phage growth requires either phage-encoded gam or host recombination functions to produce multimeric DNA for packaging. While EMBL phage libraries typically use recA+ strains, this may destabilize repetitive sequences. Alternatives include recBC- hosts, providing gam via helper plasmids in RecA- hosts, or using gam replacement vectors like Charon 34 and 35, though the latter loses genetic selection for chimeric molecules.
There is, however, a disadvantage associated with this powerful selection system. In the absence of the phage red and gam genes, phage growth requires the host recombination function ….
Lambda DNA replication shifts from theta to rolling circle mode upon ExoY10 inactivation by phage gam, which is replaced by DNA insert. Spi- phage growth now relies on host RecA for Chi-dependent oligomeric circle formation via recombination [2]. EMBL phage libraries typically use recA+ strains, potentially increasing the instability of repetitive sequences. Alternative approaches include using recBC- hosts, providing gam via helper plasmids in RecA- hosts, or using gam replacement vectors like Charon 34 and 35.
References
- Frischauf, A., Lehrach, H., Poustka, A., & Murray, N. (1983). Lambda replacement vectors carrying polylinker sequences. Journal of Molecular Biology, 170(4), 827–842. https://doi.org/10.1016/s0022-2836(83)80190-9.
- Frischauf, A., Murray, N., & Lehrach, H. (1989). λ Phage Vectors—EMBL Series. In Elsevier eBooks (pp. 295–307). https://doi.org/10.1016/b978-0-12-765560-4.50022-8
- Karn, J., Brenner, S., & Barnett, L. (1983). [1] New bacteriophage lambda vectors with positive selection for cloned insects. Methods in Enzymology on CD-ROM/Methods in Enzymology, 3–19. https://doi.org/10.1016/0076-6879(83)01004-6
- FREDERICQ, P. “Transfert génétique des propriétés lysogènes chez E. coli” [Genetic transfer of lysogenic property in E. coli]. Comptes rendus des seances de la Societe de biologie et de ses filiales vol. 147,23-24 (1953): 2046-9.
- Agilent Technologies. (2008). Lambda EMBL3/BamH I Vector Kit. INSTRUCTION MANUAL. https://www.chem-agilent.com/pdf/strata/241211.pdf


