Next-generation sequencing (NGS) is a high-throughput sequencing method that enables sequence profiling of everything from genomes and transcriptomes to DNA-protein interactions. This article provides an overview of the NGS workflow, various NGS methods, and applications of each method.
The need for NGS technologies
Next-generation sequencing (NGS) technologies have become indispensable in genomics research and various fields due to several key advantages over traditional sequencing methods like Sanger sequencing. Here are some of the critical needs that NGS technologies address:
- Direct sequencing method
- high-throughput, accurate, and reproducible
- cost-effective
- Advances in Epigenetics
- Applications in Transcriptomics and Proteomics
Generations of Sequencing Technologies.

Maxam-Gilbert method and Sanger sequencing
DNA sequencing evolved through two main approaches. The Maxam-Gilbert method uses chemical modification and subsequent DNA backbone cleavage at modified nucleotides, though its use of toxic reagents limited adoption. The preferred Sanger sequencing method employs dideoxynucleotides (ddNTPs) lacking 3′-OH groups, which terminate DNA chain elongation upon incorporation. These ddNTPs, tagged with radioactive or fluorescent labels for detection, enable automated sequencing. Sanger’s sequencing by synthesis (SBS) approach has become the standard due to its reliability and safer chemistry (Slatko et al., 2018).
Differences between NGS and Sanger sequencing
| Characteristics | Sanger Sequencing | Next-Generation Sequencing (NGS) |
|---|---|---|
| Time | Rapid results obtainable within 30 minutes | Flexible with high throughput; complete analysis within 3 hours |
| Workflow | Single run performs both Sanger sequencing and fragment analysis | Automated operation from DNA to data analysis with minimal manual intervention |
| Advantages | Rapid and cost-effective for small target sets (1-20 targets) | Higher sequencing depth with improved sensitivity (≤1%); Enhanced discovery potential; Superior mutation resolution; Greater data output from equivalent DNA input; Increased sample throughput |
How DNA sequencing with next-generation sequencing (NGS) works
Next-generation sequencing (NGS) is a revolutionary technology that allows for the rapid sequencing of DNA and RNA. The NGS workflow can be divided into several key and basic steps:
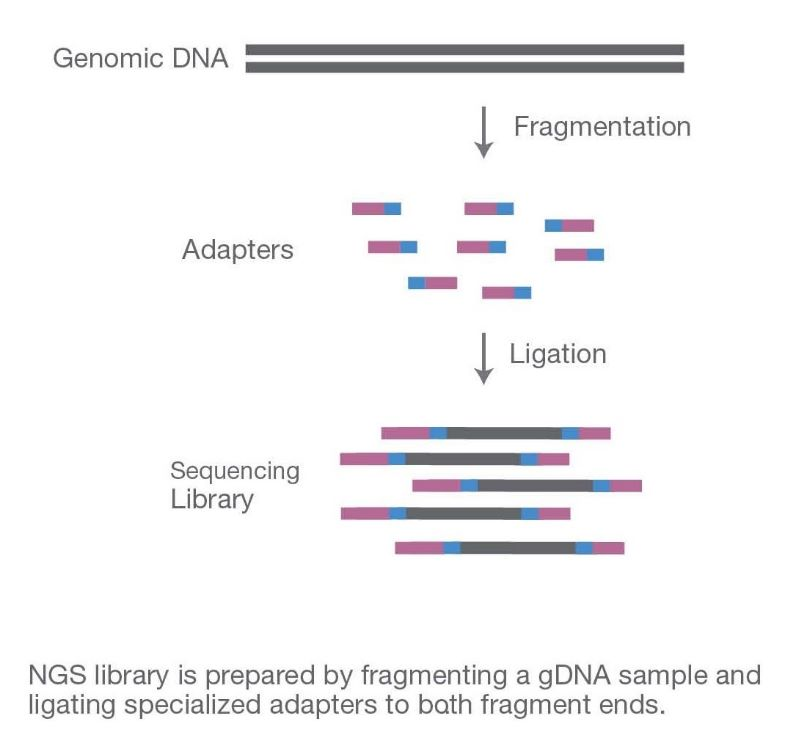
Sample Preparation
DNA or RNA is extracted from biological samples, such as blood, tissue, or cells. Extracted nucleic acids are fragmented, and specific adapters are ligated to the ends of these fragments to create a library. This step ensures that the fragments can be efficiently sequenced.
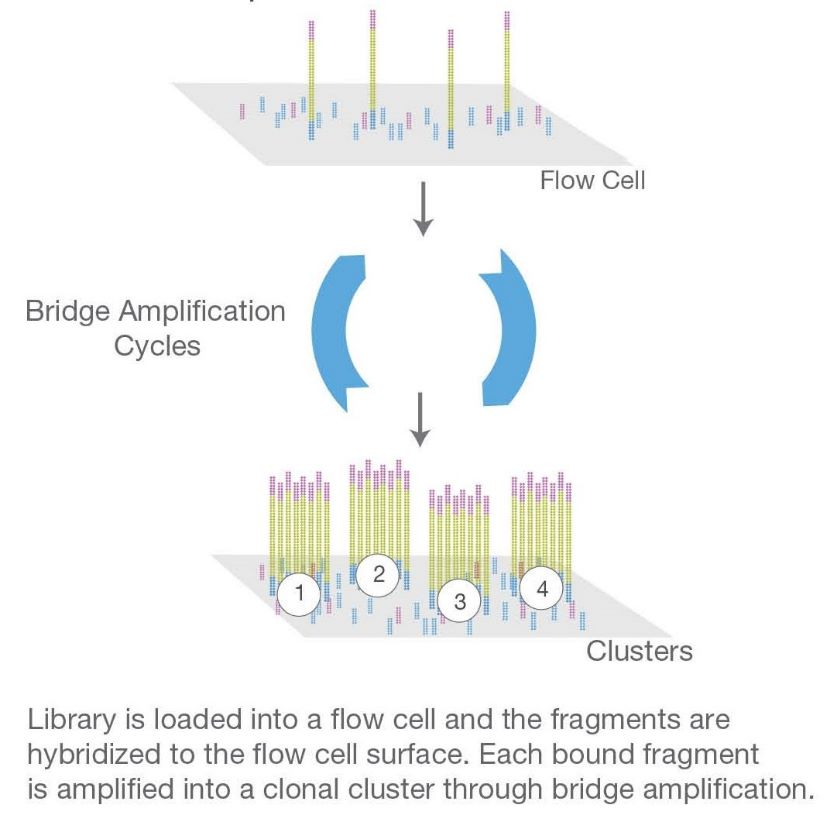
Amplification
The prepared library is then amplified using polymerase chain reaction (PCR) to increase the quantity of DNA available for sequencing. This step enriches the library for the target sequences.
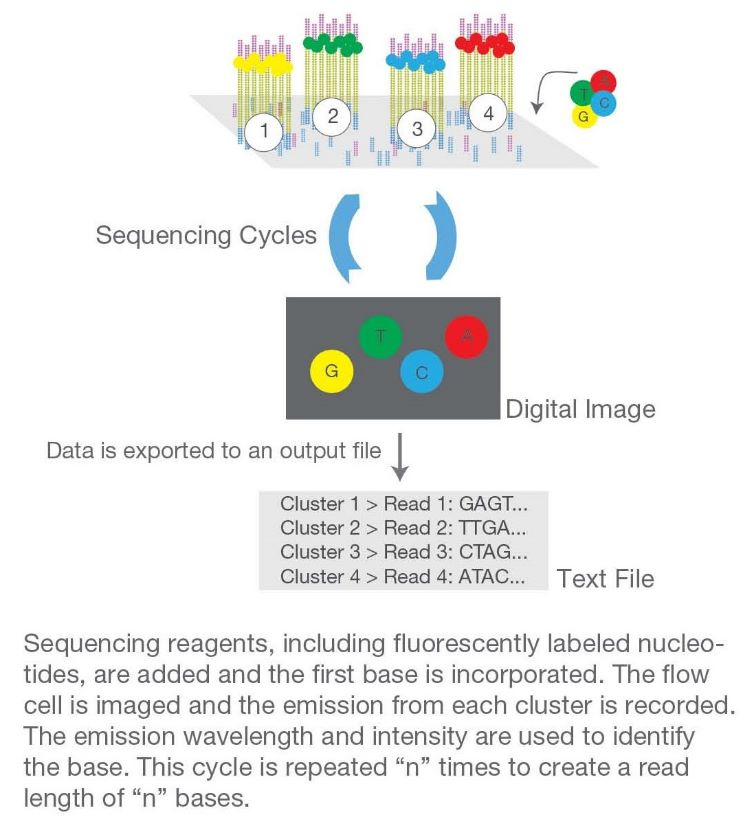
Sequencing
The amplified library is loaded onto a flow cell, where clonal amplification occurs, generating millions of dense clusters of identical DNA fragments. The sequencing process involves the incorporation of fluorescently labeled nucleotides, where each incorporated base emits a specific signal. This process is repeated to read the sequence of bases in each cluster.
Data Analysis
The fluorescent signals are converted into sequence data (raw reads) using software algorithms. The raw data undergoes quality control to assess the accuracy and reliability of the sequencing results. The processed reads are aligned to a reference genome, and variations such as single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) are identified.
Interpretation and Reporting
Advanced bioinformatics tools are used to interpret the sequencing data, providing insights into genomic variations and their potential biological implications. The findings are compiled into a report, detailing the results of the sequencing analysis, which can be used for clinical diagnosis, research, or further investigations.
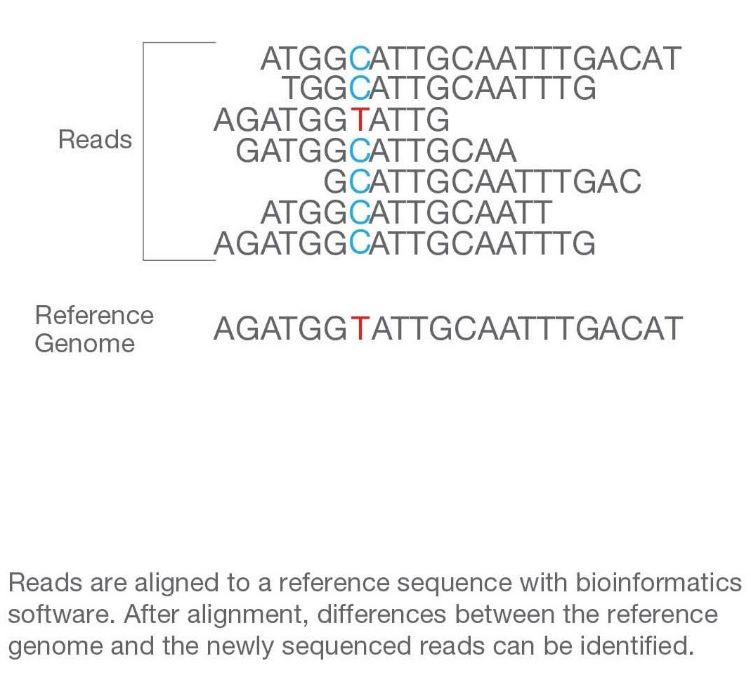
What are the types of NGS?
| Platform | Sequencing Technology | Amplification Type | Read Length (bp) |
|---|---|---|---|
| 454 pyrosequencing | Sequencing by synthesis | Emulsion PCR | 400–1000 |
| Ion Torrent | Sequencing by synthesis | Emulsion PCR | 200–400 |
| Illumina | Sequencing by synthesis | Bridge PCR | 36–300 |
| SOLiD | Sequencing by ligation | Emulsion PCR | 75 |
| DNA nanoball sequencing | Sequencing by ligation | Amplification by Nanoball PCR | 50–150 |
| Helicos single-molecule sequencing | Sequencing by synthesis | Without Amplification | 35 |
| PacBio Onso system | Sequencing by binding | Optional PCR | 100–200 |
| Platform | Sequencing Technology | Read Length (bp) |
|---|---|---|
| PacBio Single-molecule real-time sequencing (SMRT) technology | Seq by synthesis | 10,000–25,000 |
| Nanopore DNA sequencing | Sequence detection through electrical impedance | 10,000–30,000 |
| Platform | Principle | Limitations |
|---|---|---|
| 454 pyrosequencing | Detection of pyrophosphate released during nucleotide incorporation. | Homopolymer errors; |
| Ion Torrent | Ion semiconductor sequencing principle detecting H+ ion generated during nucleotide incorporation. | Homopolymer errors; Lower throughput than Illumina |
| Illumina | Solid-phase sequencing on immobilized surface leveraging clonal array formation using proprietary reversible terminator technology for rapid and accurate large-scale sequencing using single labeled dNTPs, which are added to the nucleic acid chain. | Short read lengths; GC bias; Difficulty with repetitive regions |
| SOLiD | Enzymatic method of sequencing using DNA ligase. 8-Mer probes with a hydroxyl group at 3′ end and a fluorescent tag (unique to each base A, T, G, C) at 5′ end are used in the ligation reaction. | GC bias; Short reads. |
| DNA nanoball sequencing | Splint oligo hybridization with post-PCR amplicon from libraries forms circles. Circular ssDNA is used as the DNA template to generate long strings that self-assemble into DNA nanoballs. These are bound to amino silane-coated flow cells. | Multiple PCR cycles and exhaustive workflow limit efficiency. |
| Helicos single-molecule sequencing | Poly-A-tailed short 100–200 bp fragmented genomic DNA sequenced on poly-T oligo-coated flow cells using fluorescently labeled 4 dNTPs. The signal released upon nucleotide addition is captured. | Highly sensitive instrumentation is required. As sequence length increases, fewer strands can be utilized. |
| PacBio Onso system | Sequencing by binding (SBB) chemistry uses native nucleotides and scarless incorporation under optimized conditions for binding and extension. | Higher cost compared to other platforms. |
| PacBio Single-molecule real-time sequencing (SMRT) technology | SMRT sequencing employs SMRT Cell housing zero-mode waveguides (ZMWs). DNA molecules immobilized in ZMWs emit light during polymerase activity, allowing real-time nucleotide incorporation. | Higher cost per base; Lower throughput; Higher raw error rate |
| Nanopore DNA sequencing | DNA or RNA molecules pass through biological nanopores (8 nm wide). Electrophoretic mobility generates a current signal as linear nucleic acid strands pass through. | Higher error rate; Lower throughput; DNA quality dependent |
Platforms for next-generation sequencing and their applications
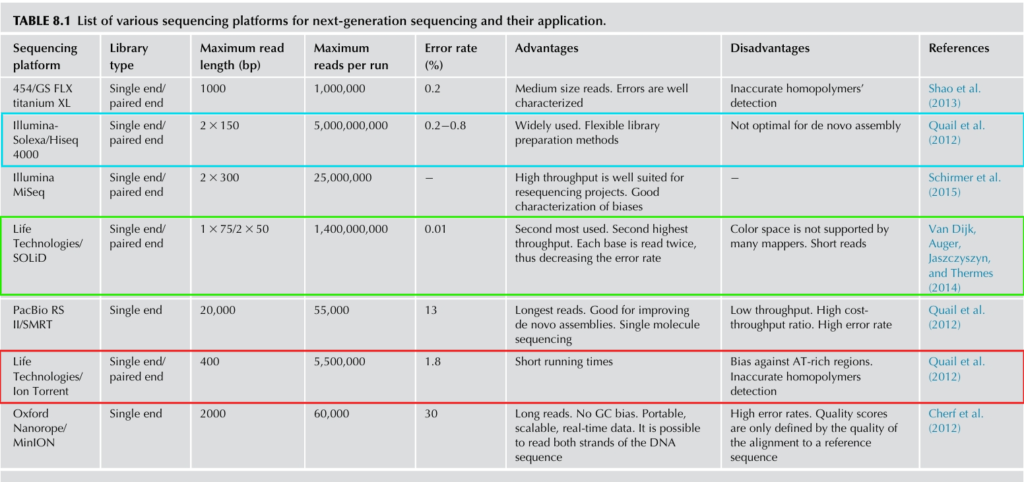
NGS applications and their preferred sequencing types.
| Research Area | Application | Preferred NGS Type(s) | Rationale for Platform Choice |
|---|---|---|---|
| Genomics | |||
| Whole Genome Sequencing | De novo assembly; Variant detection; Structural analysis | Illumina (primary); PacBio/Nanopore (complex regions) | High accuracy needed for variants; Long reads for structural variants; Cost-effectiveness for large genomes |
| Targeted Sequencing | Gene panels; Mutation hotspots; Clinical diagnostics | Illumina; Ion Torrent | High accuracy; Cost-effective for small regions; Rapid turnaround |
| Transcriptomics | |||
| RNA-Seq | Gene expression; Alternative splicing; Novel transcript discovery | Illumina (quantification); PacBio (isoforms) | High throughput for expression; Full-length transcripts for isoforms |
| Small RNA Analysis | miRNA profiling; piRNA studies; siRNA analysis | Illumina | Short read length ideal for small RNAs; High accuracy for quantification |
| Epigenomics | |||
| DNA Methylation | Bisulfite sequencing; Methylation patterns | Illumina; PacBio (native detection) | High coverage needed; Direct detection capability |
| ChIP-Seq | Protein-DNA interactions; Histone modifications | Illumina | Short reads sufficient; High throughput needed |
| Metagenomics | |||
| Microbiome Studies | Species identification; Community analysis | Illumina (profiling); PacBio/Nanopore (full genomes) | High throughput for diversity; Long reads for complete assembly |
| Pathogen Detection | Clinical diagnostics; Outbreak surveillance | Nanopore; Illumina | Real-time detection capability; High accuracy for variants |
| Clinical Applications | |||
| Cancer Genomics | Somatic mutations; Tumor evolution; Drug resistance | Illumina (primary); PacBio (fusion genes) | High sensitivity for rare variants; Structural variant detection |
| Genetic Disease | Variant detection; Inheritance patterns | Illumina; 10x Genomics (phasing) | High accuracy; Haplotype information |
| Multi-omics | |||
| Integrated Studies | Combined DNA/RNA analysis; Proteogenomics | Multiple platforms | Different requirements for each assay; Complementary approaches |
| Single-Cell Analysis | Cell heterogeneity; Developmental studies | Illumina; 10x Genomics | High throughput; Cellular resolution |
References
- Satam, Heena, et al. “Next-Generation Sequencing Technology: Current Trends and Advancements.” Biology, vol. 12, no. 7, 2023, p. 997, https://doi.org/10.3390/biology12070997. Accessed 27 Oct. 2024.
- Next-Generation Sequencing (NGS) | Explore the technology. (n.d.).
- Slatko, B. E., Gardner, A. F., & Ausubel, F. M. (2018). Overview of Next‐Generation sequencing Technologies. Current Protocols in Molecular Biology, 122(1). https://doi.org/10.1002/cpmb.59


